Knockout (KO) mice play a crucial role in elucidating the mechanisms underlying the onset of human diseases and in developing therapeutic strategies. Assistant Professor Kuno has developed web-based tools that support the generation of KO mice using genome-editing technology. By integrating these tools, he aims to facilitate the easier and more reliable creation of KO mice.

KO Mice Supporting Life Science
Knockout (KO) mice, in which specific genes are deleted, serve as a powerful tool for studying gene functions and their relationships to diseases. Although the generation of KO mice previously required considerable time and effort, this has markedly changed with the advent of genome-editing technology. KO mice are now easily produced in a short period of time, and research in this field is becoming increasingly active.
Genome editing is a technology that uses DNA-cutting enzymes to cleave or modify specific target sequences in the genome, with CRISPR-Cas9 being the most representative method.
Before genome editing, a precise design process is required to identify which regions to cut. If this design is flawed, the target gene cannot be successfully knocked out. After genome editing, a genotyping analysis is necessary to confirm whether the intended mutations have occurred according to the design. However, this step has posed many challenges that are unique to genome editing and has required considerable effort.
Although the production of KO mice has been simplified, significant challenges remain in the processes before and after genome editing.
Supporting KO Mouse Production “Before and After” Through Tool Development
I created a KO mouse for the first time during my graduate studies when I was conducting disease research using mice. At that time, I learned how to design genome-editing experiments, which required specialized knowledge and experience. I realized that it was difficult for beginners to construct high-quality designs on their own, and I also thought that it may be possible to automate much of the process using a computer.
This experience led me to develop “KOnezumi” (*1), a web-based tool that automatically generates genome-editing designs simply by entering the name of the gene to be knocked out (Figure 1).
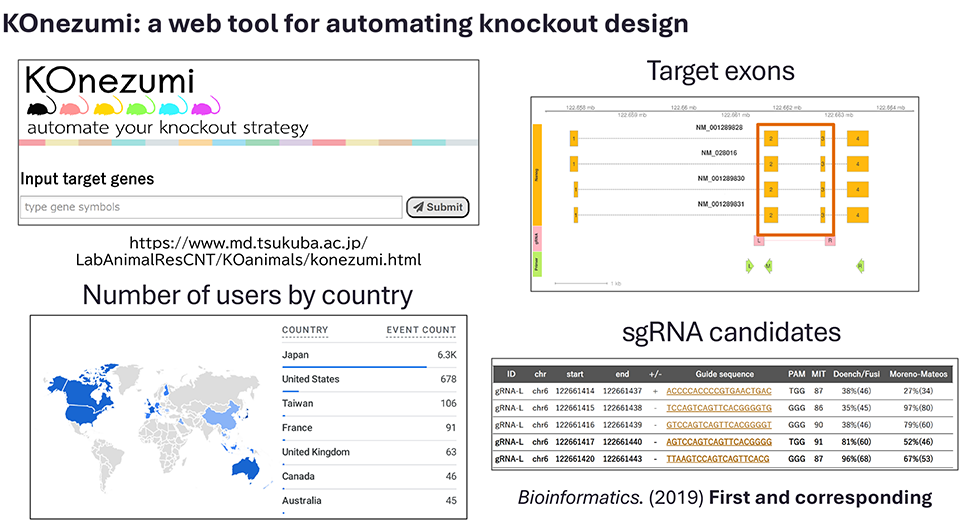
Figure 1. The fully automated KO design generation tool KOnezumi (captured from the KOnezumi website)
When I generated mice, I noted that many of the offspring carried various “unintended mutations” that differed from the designed mutations. Conventional genotyping techniques made it difficult to accurately identify these unintended mutations. To address this, I developed “DAJIN” (*2), a tool that comprehensively analyzes the outcomes of genome editing. With DAJIN, it is possible to reveal all the edits that occur in the genome regions targeted by CRISPR-Cas9 and to perform a comprehensive analysis of the mouse genome (Figure 2).

Figure 2. The genotyping analysis tool DAJIN, which integrates nanopore sequencing and deep learning
The figure on the left was created by Assistant Professor Kuno. The figure on the right was modified from https://doi.org/10.1371/journal.pbio.3001507. The sequencer illustration in the figure was modified from https://doi.org/10.7875/togopic.2017.35 ©2016 DBCLS TogoTV.
Understanding Gene Networks from Phenotypes at the Individual Level
“KOnezumi” and “DAJIN” are tools that support the before and after stages of genome editing; therefore, I thought that it may also be valuable to have a tool that supports an even earlier stage, the hypothesis generation phase, thereby helping researchers to identify “which genes or groups of genes may be interesting to target”. This idea led to the development of “TSUMUGI” (*3), a tool currently under development. TSUMUGI utilizes the vast collection of comprehensive KO mouse phenotype data accumulated to date to identify groups of genes associated with specific phenotypes. Biological phenomena and diseases are, in most cases, not caused by a single gene, but by complex mechanisms involving multiple genes that interact with one another. Our aim is to unravel the gene networks that give rise to complex phenotypes through TSUMUGI.
We believe that by advancing mouse-based research using our suite of tools, many novel insights into gene functions will be gained at the individual level. In the future, we hope to leverage these extensive datasets in order to systematically elucidate the relationships among genes involved in abnormal phenotypes, thereby contributing to a more detailed understanding of diseases as well as advances in medicine.
(Date of interview: August 27, 2025)
- (※1) https://www.md.tsukuba.ac.jp/LabAnimalResCNT/KOanimals/konezumi.html
- (※2) https://github.com/akikuno/DAJIN2
- (※3) https://larc-tsukuba.github.io/tsumugi/
All of the publicly released tools are available free of charge for anyone to use.






